import jax
from jax import random, Array
import jax.numpy as jnp
import matplotlib.pyplot as plt
from tqdm.notebook import tqdm
from typing import *
from gp import k, mk_cov, cholesky_inv, create_toy_sparse_dataset, PlotContext
key = random.PRNGKey(42) # Create a random key
KRR with Tikhonov Regularization#
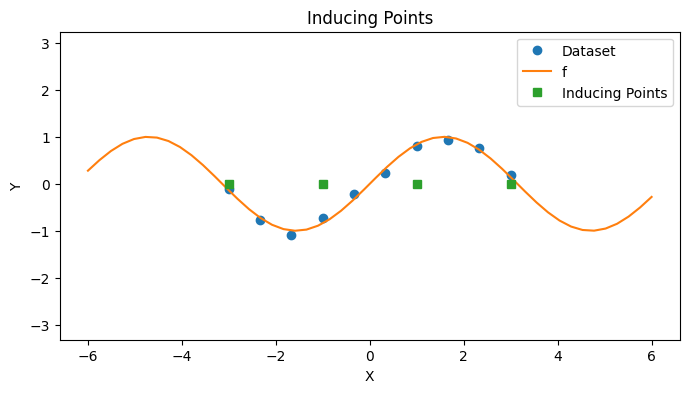
In this notebook, we’ll introduce KRR with Tikhonov regularization. Similar to how a KRR was related to a GP regression, a KRR with Tikhonov regularization will be related to the SoR sparse GP model. We reintroduce our toy dataset from before below.
# Create dataset
f = lambda x: jnp.sin(x)
xs, zs, ys, Sigma = create_toy_sparse_dataset(f, key)
N, M = len(xs), len(zs)
# Plot
with PlotContext(title="Inducing Points", xlabel="X", ylabel="Y") as ax:
plt.plot(xs, ys, marker="o", linestyle="None", label="Dataset")
plt.plot(jnp.linspace(-6, 6), f(jnp.linspace(-6, 6)), label="f")
plt.plot(zs, jnp.zeros_like(zs), marker="s", linestyle="None", label="Inducing Points")
KRR with Tikhonov Regularization#
Like a sparse GP based on inducing points, a KRR with Tikhonov regularization is parameterized by \(M\) inducing points \(z = (z_1 \dots z_m)^T\). The predictive function is given as
where
\(K_{x_* z} = (k(x_*, z_1) \dots k(x_*, z_M))^T\) is the kernel between an input and an inducing point and
\(\alpha\) is a vector of \(M\) weights that is learned. For comparison, an ordinary KRR has predictive function \(krr(x_*; \alpha) = K_{*x} \alpha\) where \(x = (x_1 \dots x_N)^T\) are the inputs in the dataset.
def krr(k: Callable, zs: Array, alpha: Array, x_star: Array) -> Array:
"""Kernel ridge regression with Tikhonov regularization
"""
K_star_u = mk_cov(k, x_star, zs).reshape(-1)
return K_star_u @ alpha
Learning the Weights: Loss Function#
Given a dataset \(\mathcal{D} = \{(x_i, y_i) \}_{1 \leq i \leq N}\), the weights of a KRR \(\alpha\) with Tikhonov regularization are choosen to minimize the loss function
(see Appendix: Checking Loss Minimization). The term \(\alpha^T K_{uu} \alpha\) performs Tikhonov regularization.
- The weights are given by are given by
and can be obtained by minimizing $\mathcal{L}(\alpha; \mathcal{D})$ with respect to $\alpha$ (See Appendix: KRR with Tikhonov Regularization).
- Note that this agrees with the mean of the test point prediction from a probabilistic view of a GP since the prediction
is the mean of $p(f_* | y)$.
- The term $\alpha^T K_{uu} \alpha$ is a **regularization term** that performs **$\ell^2$ regularization**.
- The weights correspond to a **weight-space view** of GPs.
def loss_tk(k: Callable, Sigma: Array, xs: Array, ys: Array, zs: Array, K_uu: Array, K_uf: Array, alpha: Array) -> float:
"""Loss function.
"""
# diff = ys.flatten() - K_uf.transpose() @ alpha
diff = ys.flatten() - jax.vmap(lambda x: krr(k, zs, alpha, x))(xs)
return diff.transpose() @ cholesky_inv(Sigma) @ diff + alpha.transpose() @ K_uu @ alpha
Learning the Weights: Solution#
The minimum can be solved in closed form and is given by
and can be obtained by minimizing \(\mathcal{L}(\alpha; \mathcal{D})\) with respect to \(\alpha\) (See Appendix: KRR with Tikhonov Regularization). Note that this agrees with the mean of the test point prediction from a SoR GP since the prediction
is the mean of \(q_{SoR}(f_* | y)\).
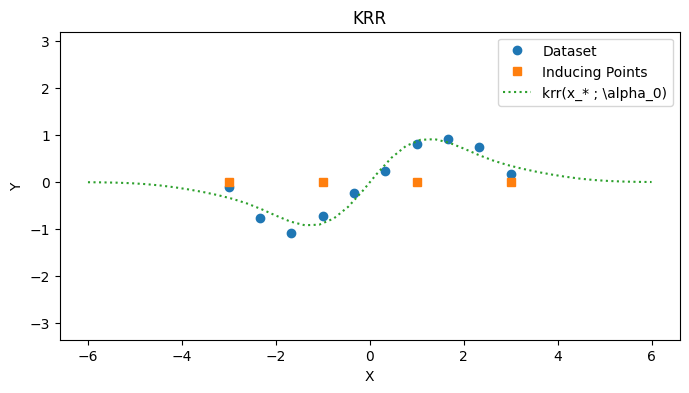
# Calculate solution
K_uu = mk_cov(k, zs, zs)
K_uf = mk_cov(k, zs, xs)
Sigma_inv = cholesky_inv(Sigma)
alpha_star = cholesky_inv(K_uu + K_uf @ Sigma_inv @ K_uf.transpose()) @ K_uf @ Sigma_inv @ ys
with PlotContext(title="KRR", xlabel="X", ylabel="Y") as ax:
# Plot dataset
plt.plot(xs, ys, marker="o", linestyle="None", label="Dataset")
plt.plot(zs, jnp.zeros_like(zs), marker="s", linestyle="None", label="Inducing Points")
# Plot KRR
test_xs = jnp.linspace(-6, 6, 40).reshape(-1, 1)
plt.plot(test_xs, [krr(k, zs, alpha_star, x) for x in test_xs], label=r"krr(x_* ; \alpha_0)", linestyle="dotted")
Tikhonov regularization reduces to KRR when x = z#
Suppose \(K = K_{uu} = K_{uf} = K_{fu} = K_{ff}\). Then
This agrees with standard GP prediction.
Summary#
Introduced KRR with Tikhonov regularization, a closely related model to SoR GP regression.
Illustrated KRR with Tikhonov regularization on a toy dataset.
Appendix: KRR with Tikhonov Regularization#
We can check that the minima of
with respect to the KRR model is
We can compute
Appendix: KRR via Loss Minimization#
def grad_desc_tk(loss: Callable, k: Array, Sigma: Array, xs: Array, ys: Array,
zs: Array, K_uu: Array, K_uf: Array, alpha_0, lr=0.00001, iterations=100):
"""Simple gradient descent procedure to optimize loss function.
"""
pbar = tqdm(range(iterations))
losses = []
for i in pbar:
step = lr * jax.jacrev(loss, argnums=7)(k, Sigma, xs, ys, zs, K_uu, K_uf, alpha_0)
alpha_0 = alpha_0 - step
l = loss(k, Sigma, xs, ys, zs, K_uu, K_uf, alpha_0)
losses += [l]
pbar.set_postfix(loss=l)
return alpha_0, losses
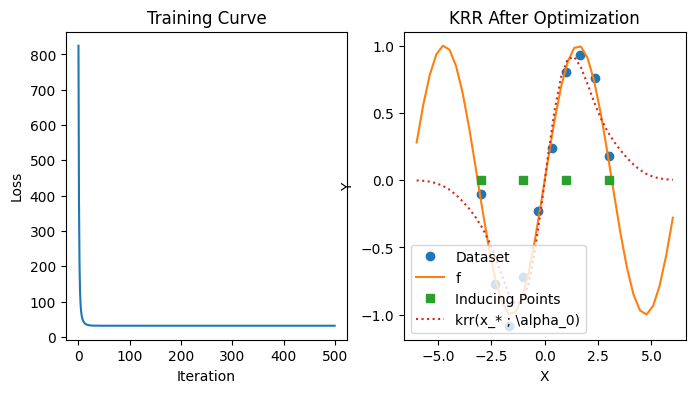
iterations = 500
alpha_star, losses = grad_desc_tk(loss_tk, k, Sigma, xs, ys, zs, K_uu, K_uf, jnp.ones(M), lr=0.0005, iterations=iterations)
plt.figure(figsize=(8, 4))
plt.subplot(1, 2, 1)
plt.plot(range(iterations), losses)
plt.title("Training Curve"); plt.xlabel("Iteration"); plt.ylabel("Loss")
plt.subplot(1, 2, 2)
plt.plot(xs, ys, marker="o", linestyle="None", label="Dataset")
plt.plot(test_xs, [f(x) for x in test_xs], label="f")
plt.plot(zs, jnp.zeros_like(zs), marker="s", linestyle="None", label="Inducing Points")
plt.plot(test_xs, [krr(k, zs, alpha_star, x) for x in test_xs], label=r"krr(x_* ; \alpha_0)", linestyle="dotted")
plt.title("KRR After Optimization"); plt.xlabel("X"); plt.ylabel("Y"); plt.legend();