import jax
from jax import random, Array
import jax.numpy as jnp
import matplotlib.pyplot as plt
from typing import *
from gp import k, mk_cov, cholesky_inv, create_toy_sparse_dataset, PlotContext
key = random.PRNGKey(42) # Create a random key
Variational Gaussian Processes (VGPs)#
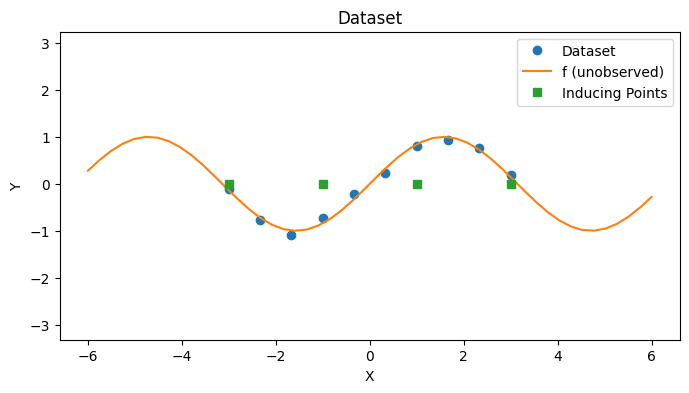
In this notebook, we explore a Variational GP (VGP) which is a kind of sparse GP aimed at solving the scaling problems of GPs by approximating the GP posterior with variational inference. As before, we’ll reintroduce a toy dataset for illustrative purposes.
# Create dataset
f = lambda x: jnp.sin(x)
xs, zs, ys, Sigma = create_toy_sparse_dataset(f, key)
N, M = len(xs), len(zs)
with PlotContext(title="Dataset", xlabel="X", ylabel="Y") as ax:
# Plot dataset and inducing points
plt.plot(xs, ys, marker="o", linestyle="None", label="Dataset")
plt.plot(jnp.linspace(-6, 6), f(jnp.linspace(-6, 6)), label="f (unobserved)")
plt.plot(zs, jnp.zeros_like(zs), marker="s", linestyle="None", label="Inducing Points")
VGP Model#
In variational inference, we approximate a complex probability distribution \(p\) with a simpler distribution \(q\) from a variational family that has more tractable properties compared to \(p\). In VGP [1], we choose the distribution \(q(f, u)\) to be a approximate the posterior \(p(f, u | y)\) where \(u\) are the evaluations of a function choosen at \(M\) inducing points \(\{ z_i \}_{1 \leq i \leq M}\). That is, we will solve
to obtain a \(q(f, u) \approx p(f, u | y)\). Posterior predictive inference can then be performed by using the posterior predictive \(q(f_* | y)\).
VGP Approximation#
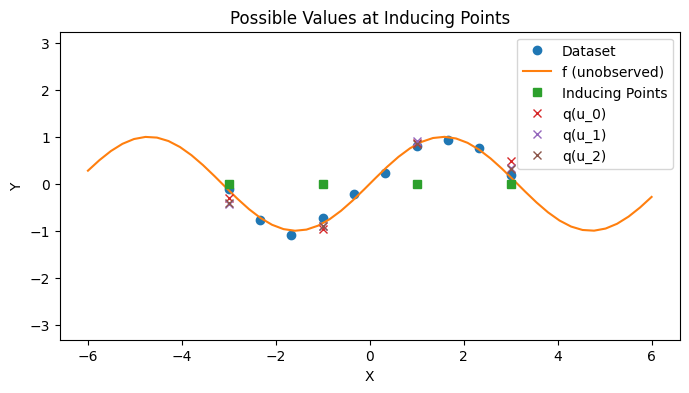
To construct the approximation \(q(f, u)\), we choose
and solve for \(q(u)\) (See Appendix for derivation). This differs from the approach taken in SoR, which assumes that \(q(u) \sim \mathcal{N}(0, K_{uu})\) and approximates \(p(f | u) \approx q_{SoR}(f | u)\). Titsias shows that
where \(C = K_{zz} + K_{zx}\Sigma^{-1} K_{xz}\).
# Compute kernel
K_zz = mk_cov(k, zs, zs)
K_zx = mk_cov(k, zs, xs)
K_xz = K_zx.transpose()
# Compute mean and covariance
Sigma_inv = cholesky_inv(Sigma)
C = K_zz + K_zx @ Sigma_inv @ K_xz
C_inv = cholesky_inv(C)
m = (K_zz @ C_inv @ K_zx @ Sigma_inv @ ys).reshape(-1)
cov = K_zz @ C_inv @ K_zz
# Sample q(u)
us = random.multivariate_normal(key, mean=m, cov=cov, shape=(3,)).transpose()
with PlotContext(title="Possible Values at Inducing Points", xlabel="X", ylabel="Y") as ax:
# Plot dataset and inducing points
plt.plot(xs, ys, marker="o", linestyle="None", label="Dataset")
plt.plot(jnp.linspace(-6, 6), f(jnp.linspace(-6, 6)), label="f (unobserved)")
# Plot q(u)
plt.plot(zs, jnp.zeros_like(zs), marker="s", linestyle="None", label="Inducing Points")
for i in range(3):
plt.plot(zs, us[:, i], marker="x", linestyle="None", label=f"q(u_{i})")
Fitting a VGP#
The posterior predictive of a VGP (see Titsias [2], eq 6 and also the Appendix) is
As a reminder, \(C = K_{zz} + K_{zx}\Sigma^{-1} K_{xz}\).
def fit_vgp(k: Callable, Sigma: Array, xs: Array, ys: Array, zs: Array):
# Compute covariances
K_zz = mk_cov(k, zs, zs)
K_xz = mk_cov(k, xs, zs)
K_zx = K_xz.transpose()
K_zz_inv = cholesky_inv(K_zz)
# Perform approximations
Sigma_inv = cholesky_inv(Sigma)
C = K_zz + K_zx @ Sigma_inv @ K_xz
C_inv = cholesky_inv(C)
alpha = C_inv @ K_zx @ Sigma_inv @ ys
cov = K_zz_inv - C_inv
return alpha, cov, K_zz, K_zz_inv
def vgp_post_pred_mean(k: Callable, zs: Array, alpha: Array, x_star: Array) -> Array:
"""Posterior predictive mean.
"""
K_star_z = mk_cov(k, x_star, zs).reshape(-1)
return K_star_z @ alpha
def vgp_post_pred_cov(k: Callable, zs: Array, cov: Array, x_star: Array) -> Array:
"""Posterior predictive covariance.
"""
K_star_star = mk_cov(k, x_star, x_star)
K_star_z = mk_cov(k, x_star, zs)
return K_star_star - K_star_z @ cov @ K_star_z.transpose()
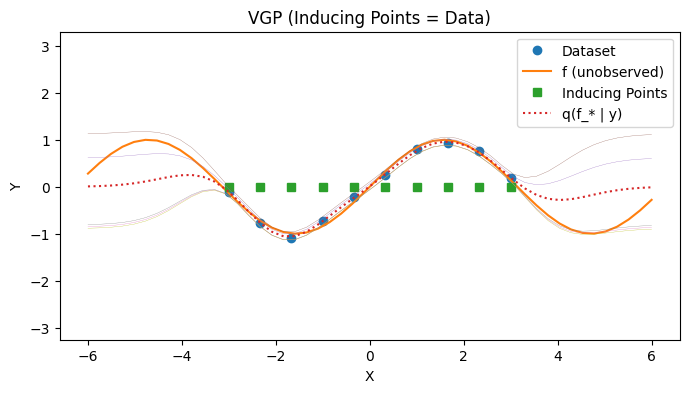
Visualizing the Posterior Predictive 1: No Inducing Points#
# Fit
zs_all_xs = xs
alpha_all_xs, cov_all_xs, K_zz_all_xs, K_zz_inv_all_xs = fit_vgp(k, Sigma, xs, ys, zs_all_xs)
# Predict
test_xs = jnp.linspace(-6, 6).reshape(-1, 1)
post_ys = [vgp_post_pred_mean(k, zs_all_xs, alpha_all_xs, x) for x in test_xs]
post_mean = jax.vmap(lambda x_star: vgp_post_pred_mean(k, zs_all_xs, alpha_all_xs, x_star))(test_xs)
post_cov = jax.vmap(lambda x_star: vgp_post_pred_cov(k, zs_all_xs, cov_all_xs, x_star), out_axes=0)(test_xs)
post_cov_ys = [random.normal(key, shape=(5,)) * jnp.sqrt(post_cov[i][0][0]) + post_mean[i] for i in range(len(test_xs))]
with PlotContext(title="VGP (Inducing Points = Data)", xlabel="X", ylabel="Y") as ax:
# Plot dataset and inducing points
plt.plot(xs[:,0], ys, marker='o', linestyle='none', label="Dataset")
plt.plot(jnp.linspace(-6, 6), f(jnp.linspace(-6, 6)), label="f (unobserved)")
plt.plot(zs_all_xs, jnp.zeros_like(zs_all_xs), marker="s", linestyle="None", label="Inducing Points")
# Plot p(f(*) | y)
plt.plot(test_xs, post_ys, label="q(f_* | y)", linestyle="dotted")
plt.plot(test_xs, post_cov_ys, linewidth=0.2)
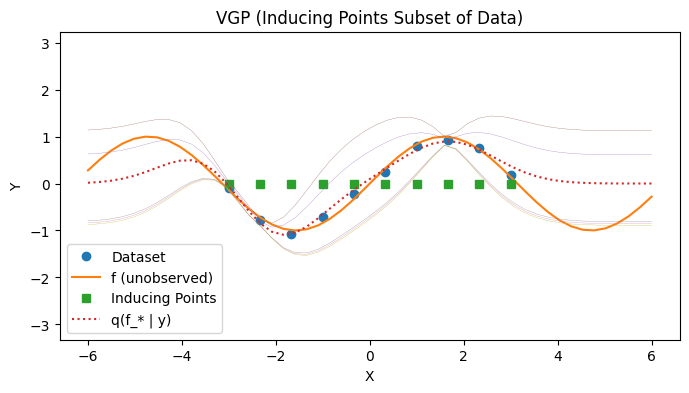
Visualizing the Posterior Predictive 2: Subset#
# Fit
zs_p = jnp.array([xs[0], xs[1], xs[-3]])
alpha_p, cov_p, K_zz_p, K_zz_inv_p = fit_vgp(k, Sigma, xs, ys, zs_p)
# Predict
test_xs = jnp.linspace(-6, 6).reshape(-1, 1)
post_ys = [vgp_post_pred_mean(k, zs_p, alpha_p, x) for x in test_xs]
post_mean = jax.vmap(lambda x_star: vgp_post_pred_mean(k, zs_p, alpha_p, x_star))(test_xs)
post_cov = jax.vmap(lambda x_star: vgp_post_pred_cov(k, zs_p, cov_p, x_star), out_axes=0)(test_xs)
post_cov_ys = [random.normal(key, shape=(5,)) * jnp.sqrt(post_cov[i][0][0]) + post_mean[i] for i in range(len(test_xs))]
# Plot
with PlotContext(title="VGP (Inducing Points Subset of Data)", xlabel="X", ylabel="Y") as ax:
# Plot dataset and inducing points
plt.plot(xs[:,0], ys, marker='o', linestyle='none', label="Dataset")
plt.plot(jnp.linspace(-6, 6), f(jnp.linspace(-6, 6)), label="f (unobserved)")
plt.plot(zs_all_xs, jnp.zeros_like(zs_all_xs), marker="s", linestyle="None", label="Inducing Points")
# Plot p(f(*) | y)
plt.plot(test_xs, post_ys, label="q(f_* | y)", linestyle="dotted")
plt.plot(test_xs, post_cov_ys, linewidth=0.2)
Summary#
We have seen how variational inference can be applied to make GP inference tractable.
We have illustrated VGP on a toy dataset.
References#
Appendix#
Appendix: VGP Derivation#
We want to solve
where
Titsias shows that
where \(C = K_{zz} + K_{zx}\Sigma^{-1} K_{xz}\).
Generative Process#
As a reminder, the generative process for a GP with inducing points is
where \(\Sigma = \beta^2I\) and \(Q_{xx} = K_{xz}K_{zz}^{-1}K_{zx}\). We will abbreviate \(u(z) = u\) and \(f(x) = f\).
Note that
VGP Derivation: ELBO#
Titsias shows in Appendix A [1] that
VGP Derivation: ELBO Inner Integral#
We solve for the inner integral. Titsias further shows in Appendix A [1] that
where \(\alpha = K_{xz}K_{zz}^{-1}u\) is mean of \(p(f \,|\, u)\) and \(B = K_{xx} - Q_{xx}\) is covariance of \(p(f \,|\, u)\).
VGP Derivation: Continue ELBO Simplification#
We can now continue to simplify the ELBO.
VGP Derivation: Solving ELBO for q(u) step 1#
We now need to find the optimal \(q(u)\). We can see that \(q(u) \propto \mathcal{N}(y \,|\, \alpha, \Sigma) p(u)\) by differentiating the ELBO at Eq * w.r.t. \(q(u)\) and setting it to \(0\).
VGP Derivation: Solving ELBO for q(u) step 2#
We can complete the square using
to obtain
where
Titsias further simplies this to
where \(C = K_{zz} + K_{zx}\Sigma^{-1} K_{xz}\) as required.
Appendix: Posterior Predictive#
Variational Family#
Note that
where
\(m = K_{zz} C^{-1} K_{zx}\Sigma^{-1}y\),
\(A = K_{zz} C^{-1} K_{zz}\), and
\(C = (K_{zz} + K_{zx}\Sigma^{-1}K_{xz})\).
